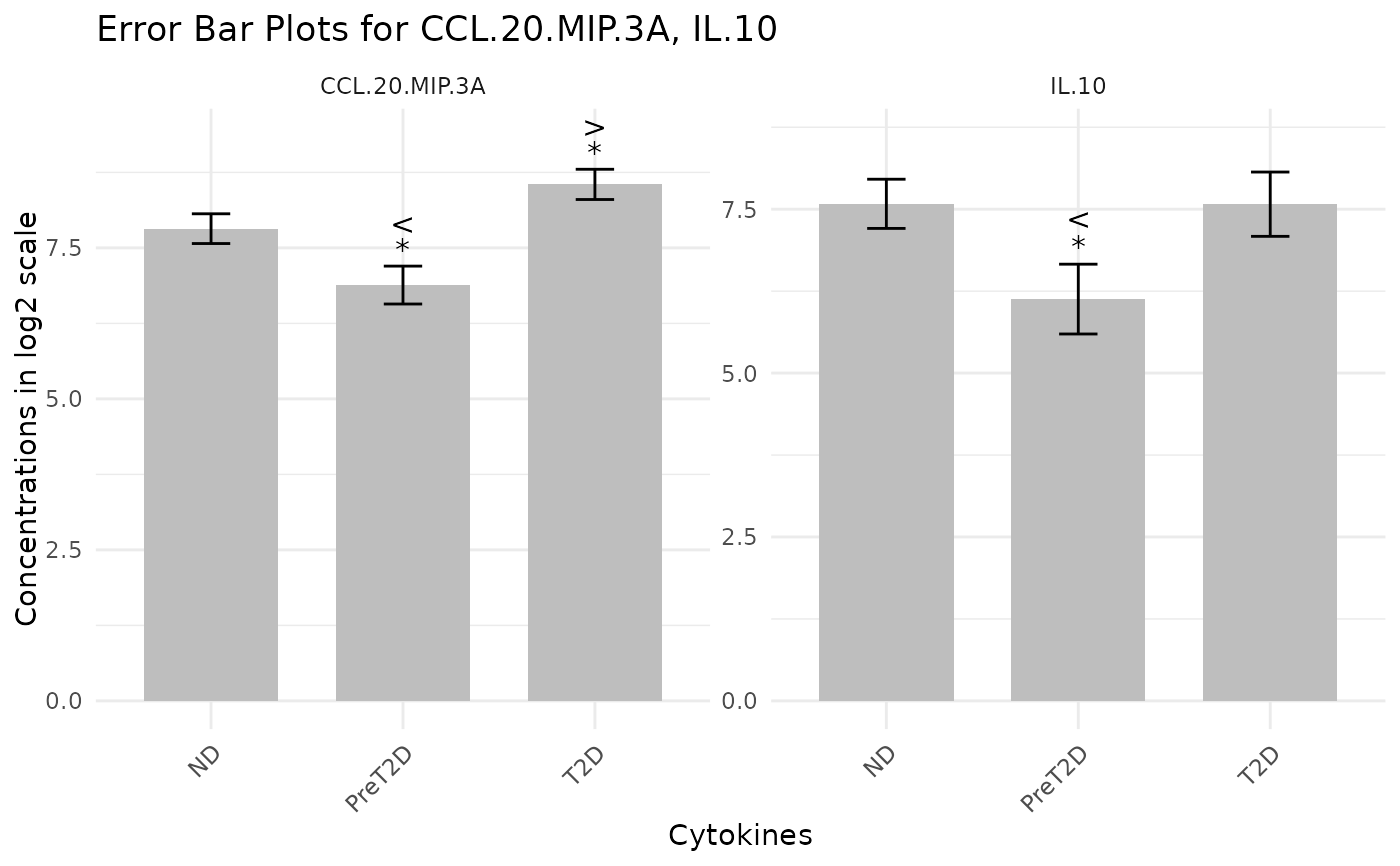
This function automatically detects a numeric measurement column from the provided data and calculates group metrics (sample size, mean, standard deviation, standard error) and performs a t-test (comparing each group to the baseline) to obtain p-values and a standardized mean difference (as a proxy for effect size). When a grouping variable is provided, it is used to separate the data into groups (the first level is taken as the baseline). If no grouping variable is provided, the metrics are computed for the overall data. The function then produces a ggplot2 bar plot with error bars and, if requested, overlays p-value and effect size annotations (displayed as symbols by default).
Usage
cyt_errbp(
data,
group_col = NULL,
p_lab = FALSE,
es_lab = FALSE,
class_symbol = TRUE,
x_lab = "",
y_lab = "",
title = NULL,
progress = NULL,
log2 = FALSE,
output_file = NULL
)Arguments
- data
A data frame containing at least one numeric variable. If a grouping variable is provided, it must be one of the columns.
- group_col
Character. (Optional) The name of the grouping (categorical) variable. If not provided, metrics are calculated for the overall data.
- p_lab
Logical. Whether to display p-value labels. Default is
FALSE.- es_lab
Logical. Whether to display effect size labels. Default is
FALSE.- class_symbol
Logical. If
TRUE, p-values and effect sizes are shown as symbols; ifFALSE, numeric values are displayed. Default isFALSE.- x_lab
Character. Label for the x-axis. Defaults to the grouping variable name if provided, or "Group" otherwise.
- y_lab
Character. Label for the y-axis. Defaults to the name of the selected numeric variable.
- title
Character. The plot title.
- progress
Optional. A Shiny
Progressobject for reporting progress updates.- log2
Logical. If
TRUE, transforms numeric variables using log2 transformation. IfFALSE, uses raw values from provided data. Default isFALSE.- output_file
Optional. A string representing the file path for the PDF file to be created. If NULL (default), the function returns a list of ggplot objects.
Examples
data <- ExampleData1
cyt_errbp(data[,c("Group", "CCL.20.MIP.3A", "IL.10")], group_col = "Group",
p_lab = TRUE, es_lab = TRUE, class_symbol = TRUE, x_lab = "Cytokines",
y_lab = "Concentrations in log2 scale", log2 = TRUE)