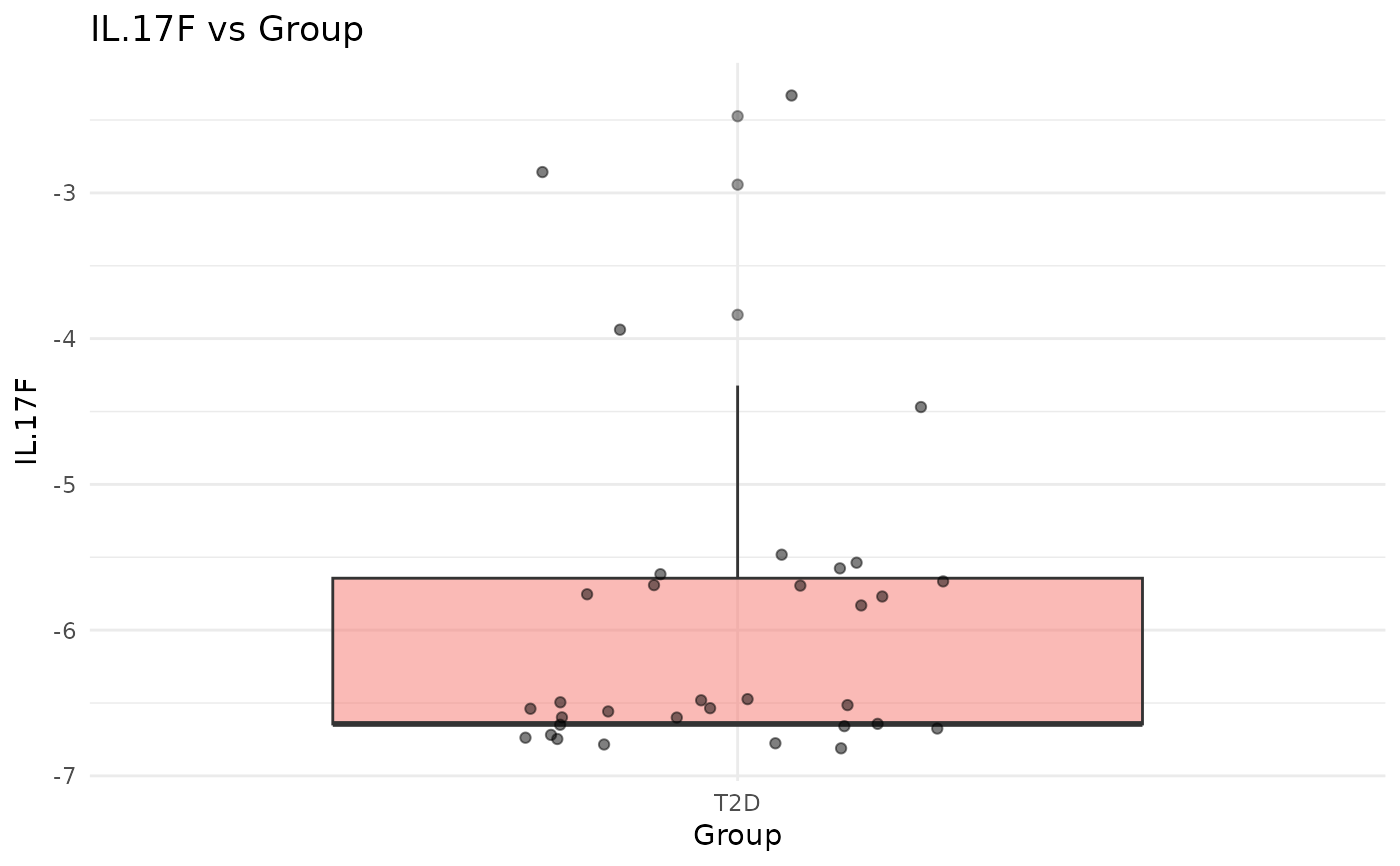
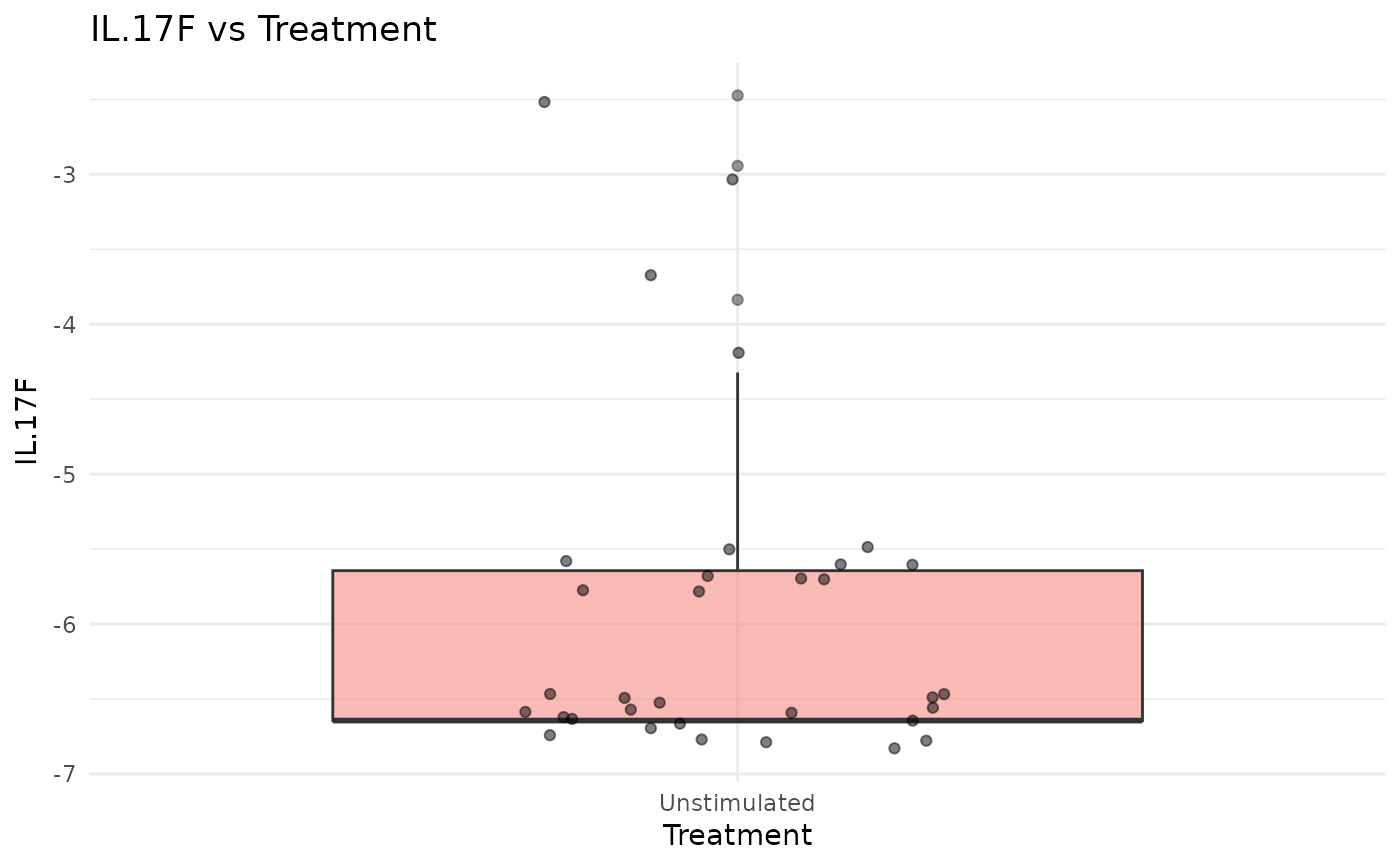
This function generates boxplots for each combination of numeric and factor variables in the provided data.
Character columns are converted to factors and the function checks that the data contains at least one numeric
and one factor column. If the scale argument is set to "log2", numeric columns are log2-transformed.
The function then creates boxplots using ggplot2 for each numeric variable grouped by each factor variable.
If output_file is provided, the plots are saved to that PDF file; otherwise, a list of ggplot objects is returned.
Usage
cyt_bp2(
data,
output_file = NULL,
mf_row = c(1, 1),
scale = NULL,
y_lim = NULL,
progress = NULL
)Arguments
- data
A matrix or data frame of raw data.
- output_file
Optional. A string representing the file path for the PDF file to be created. If NULL (default), the function returns a list of ggplot objects.
- mf_row
A numeric vector of length two specifying the layout (rows and columns) for the plots in the PDF output. Defaults to c(1, 1). (Ignored when returning ggplot objects.)
- scale
Transformation option for continuous variables. Options are NULL (default) and "log2". When set to "log2", numeric columns are transformed using the log2 function.
- y_lim
An optional numeric vector defining the y-axis limits for the plots.
- progress
Optional. A Shiny
Progressobject for reporting progress updates.